export PYTHONPATH=COSMOMC..PATH/python:$PYTHONPATHwhere COSOMC..PATH is the full path of wherever you installed CosmoMC. If you have problems on a Mac or need to install python, see Installing Python.
Scripts for plotting and analysing are described below. See also grid scripts and GetDist GUI documentation. For plotting from Planck chains see the Planck readme for how to download and install.
The getdist.plots module (directly, or via planckStyle) is used to make plots from chain results using your own custom scripts.
The sample scripts (under batch3/outputs for 2018 results, or batch2/outputs for 2015 results) make use of planckStyle. This can be interchanged with getdist.plots if you do not want to use the Planck style (or some extra functions used in the batch3/output samples). Both implement the functions
import planckStyle
g = planckStyle.getSinglePlotter(chain_dir = './PLA')
roots = ['base_nnu_mnu_plikHM_TTTEEE_lowl_lowE', 'base_nnu_mnu_plikHM_TTTEEE_lowl_lowE_post_lensing', 'base_nnu_mnu_plikHM_TTTEEE_lowl_lowE_lensing_BAO']
g.plot_2d(roots, 'mnu', 'nnu', filled=True)
g.add_legend(['Planck', 'Planck+lensing', 'Planck+lensing+BAO'], legend_loc='upper right');
g.export('mnu_nnu.png')
(for 2018 chains just change TT_lowTEB to TTTEEE_lowl_lowE)
The chain_dir argument can be neglected if you have set up a default in config.ini.
You can do "from pylab import *" and use standard matplotlib commands to customize the plots, and
there are also many plot script options you can use to customize settings, and also change global getdist plot settings using g.settings.xxx.
For example:
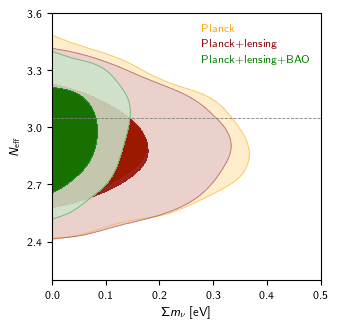
import planckStyle
g = planckStyle.getSinglePlotter(chain_dir = './PLA', ratio=1)
roots = ['base_nnu_mnu_plikHM_TTTEEE_lowl_lowE', 'base_nnu_mnu_plikHM_TTTEEE_lowl_lowE_post_lensing', 'base_nnu_mnu_plikHM_TTTEEE_lowl_lowE_lensing_BAO']
g.settings.solid_contour_palefactor = 0.8
g.plot_2d(roots, 'mnu', 'nnu', filled=True, colors=['orange', 'darkred', 'green'], lims=[0, 0.5, 2.2, 3.6])
g.add_legend(['Planck', 'Planck+lensing', 'Planck+lensing+BAO'], legend_loc='upper right', colored_text=True);
g.add_y_marker(3.046)
g.export('mnu_nnu2.png')
Outputs of the two versions should look like this: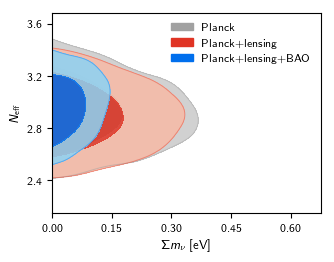
See sample scripts in batch2/outputs, the Plot Gallery and tutorial, and the full GetDist documentation. Note that if you have installed CosmoMC, you don't need to separately install the GetDist python package.
See python/getdist/chains.py and python/getdist/mcsamples.py for other functions you can use.
Use GetDist or mcsamples.py if you want to reproduce Planck results. The mcsamples module gives you code access to most getdist results, and an MCSamples instance
can be obtained from a grid as above, or you can load a chain file directly.
For example if you want to do a power law fit in the variable Ωm and H0, you could do
Samples can also be loaded directly from single chains, optionally with custom settings, e.g.
Analysis scripts
The GetDist program can be used to get means, variances, limits etc from all parameters in a chain. The python scripts allow you to do
this dynamically, and also offer additional features such as being able to define new derived parameters.
Calculating derived parameters
For simple calculations like finding the mean and variance of new derived parameters you can use the functions in python/chains.py. For example, if you want to calculate the posterior mean and limits for σ8 Ωm0.6 from Planck you could write a python script
Here p.omegam is a vector of parameter values, similar p.sigma8; samples.mean and samples.std sum the samples with the corresponding weights to calculate the result.
import getdist.plots as gplot
g = gplot.getSinglePlotter(chain_dir=r'./PLA')
samples = g.sampleAnalyser.samplesForRoot('base_plikHM_TTTEEE_lowl_lowE_lensing')
p = samples.getParams()
derived = p.sigma8 * p.omegam ** 0.6
print('mean = %s, err = %s'%(samples.mean(derived), samples.std(derived)))
print('95%% limits: %s'%samples.twoTailLimits(derived, 0.95))
which fits Ωm and H0, using log transforms (L), normalized so the exponent of Ωm is unity (as in GetDist PCA outputs).
The output includes
import GetDistPlots as s
g = s.getSinglePlotter(chain_dir=r'./PLA')
samples = g.sampleAnalyser.samplesForRoot('base_plikHM_TTTEEE_lowl_lowE_lensing')
print(samples.PCA(['omegam', 'H0'], 'LL', 'omegam'))
Principle components
PC1 (e-value: 0.008353)
[0.023238] (\Omega_m/0.315200)^{1.000000}
[0.023238] (H_0/67.357701)^{2.923650}
= 1.000005 +- 0.003003
which tells you that ΩmH02.92 is constrained at the 0.3% level.
import getdist
samples = getdist.loadMCSamples(r'./PLA/base/plikHM_TTTEEE_lowl_lowE_lensing/base_plikHM_TTTEEE_lowl_lowE_lensing', settings={'ignore_rows':0.3})
Or from a grid or list of directories you can find chains automatically e.g.
import planckStyle as s
g = s.getSinglePlotter(chain_dir=['./PLA', './mychains/xxx/'])
samples = g.getSamples('base_plikHM_TTTEEE_lowl_lowE_lensing')
p = samples.getParams()
rd_fid = 149.28
rsH = p.Hubble051 * p.rdrag / rd_fid
samples.addDerived(rsH, name='rsH', label=r'H(0.51) (r_{\mathrm{drag}}/r_{\mathrm{drag}}^{\rm fid})\, [{\rm km} \,{\rm s}^{-1}{\rm Mpc}^{-1}]')
After doing this, you can use 'rsH' as you would any of the original parameter names in the chain. If your new parameter has a hard boundary, set the
range parameter for addDerived with the [min, max] bounds.
from getdist import mcsamples samples = mcsamples.MCSamples(samples=sample_points, loglikes=loglikes, names=names)where sample_points is a matrix of sample values, names are the parameter names (list of strings), and loglikes is (optionally) an array of corresponding -log(likelihood) values.
from getdist.densities import Density2D import getdist.plots as gplot import numpy as np g = gplot.getSinglePlotter(chain_dir=r'./PLA') ... xvalues = np.arange(85, 110, 0.3) yvalues = np.arange(1000, 1500, 4) x,y = np.meshgrid(xvalues, yvalues) loglike = my_loglike_func(x,y) density = Density2D(xvalues,yvalues, np.exp(-loglike / 2)) density.contours = np.exp(-np.array([1.509, 2.4477]) ** 2 / 2) g.add_2d_contours(root, 'x', 'y', filled=True, density=density)
sudo port install python27 sudo port select --set python python27 sudo port install py-matplotlib sudo port install py-scipy sudo port install py-pyside sudo port install texlive-latex-extra sudo port install texlive-fonts-recommended sudo port install dvipng